The Language of Molecules
Understanding how molecules interact is at the heart of biology and pharmacology. Whether it is a hormone binding to its receptor, an enzyme recognizing its substrate, or a drug targeting a protein, these ligand-receptor interactions are the foundations of life and therapies. Structural bioinformatics provides us with powerful tools to visualize, model, and predict these interactions, thereby accelerating drug discovery and the understanding of biological processes.
In this article, we will explore an essential bioinformatics pipeline, using key open-access or widely used resources and software to study these fascinating interactions: PDB, BLAST, PyMOL, SWISS-MODEL, Discovery Studio, I-TASSER, and molecular docking software.
Step 1: Identify the Receptor and the Ligand – The Role of PDB and BLAST
Before diving into the interactions, it is necessary to identify the actors: the receptor (often a protein) and the ligand (a small molecule, a peptide, etc.).
PDB (Protein Data Bank): The Library of 3D Structures
The PDB is the world's primary archive of experimental structural data for biological macromolecules, determined by X-ray crystallography, NMR, or cryo-EM.
PDB: A Resource for Structural Biology
- How to use it?
- How to use it?
If the 3D structure of your receptor protein (or a homologous protein) is already known and published, you will find it here. You can search by protein name, PDB identifier, organism, etc. Often, protein structures are co-crystallized with ligands, which already provides valuable information about binding sites.
BLAST (Basic Local Alignment Search Tool): Finding Homologs
What is it?
If the 3D structure of your target receptor is not in the PDB, BLAST is your first resort. It allows you to find similar (homologous) sequences in large databases (such as GenBank or UniProt).
- How to use it?
- How to use it?
Use the peptide sequence of your receptor (blastp) to search for homologs whose 3D structure is already available in the PDB (via the PDB database on BLAST). A high sequence similarity suggests structural similarity, potentially allowing you to proceed to homology modeling.
Step 2: Model the 3D Structure of the Receptor (if necessary) – SWISS-MODEL and I-TASSER
If no suitable homologous structure is found in the PDB, or if your receptor has a specific conformation to model, you will need to resort to structure prediction.
SWISS-MODEL: Homology Modeling Made Easy

- What is it?
- What is it?
An automated web server for homology modeling. It uses a protein sequence as input and searches for homologous proteins with a known 3D structure (the "templates") in the PDB.
- How to use it?
- How to use it?
You submit the sequence of your receptor protein. SWISS-MODEL identifies the best templates and builds a 3D model based on the sequence alignment and the structure of the template. This is the preferred approach if you have a good template (high sequence identity > 30-40%).
I-TASSER: For More Complex Cases (or Without Direct Homologs)
- What is it?
- What is it?
A server for protein structure prediction using a "threading" method and ab initio simulation. It is often used when there is no obvious template with high sequence identity.
- How to use it?
- How to use it?
You submit the sequence of your protein. I-TASSER generates several possible 3D models and ranks them by confidence. It is a more complex method but potentially more accurate for "difficult" proteins.
Step 3: Visualization and Preparation – PyMOL
Once you have the 3D structure of your receptor (either from the PDB or modeled), it is crucial to visualize and prepare it.
PyMOL: Your Molecular Visualization Workshop
- What is it?
A very popular, versatile, and powerful open-source molecule viewer. It allows for the creation of high-quality 3D representations of proteins, nucleic acids, and small molecules.
- How to use it?
- How to use it?
- Load structures: Import your PDB file or the generated model.
- Explore: Switch between different representation modes (spheres, sticks, ribbons, surfaces) to visualize domains, binding cavities, and important residues.
- Identify the binding sites: Locate the hydrophobic/hydrophilic cavities, charged residues, and key amino acids for interaction.
- Prepare for docking: Clean the structure (remove water, excess ions), add hydrogens, assign atomic charges if necessary. This is the step where you "clean" your receptor to make it ready for docking simulation.
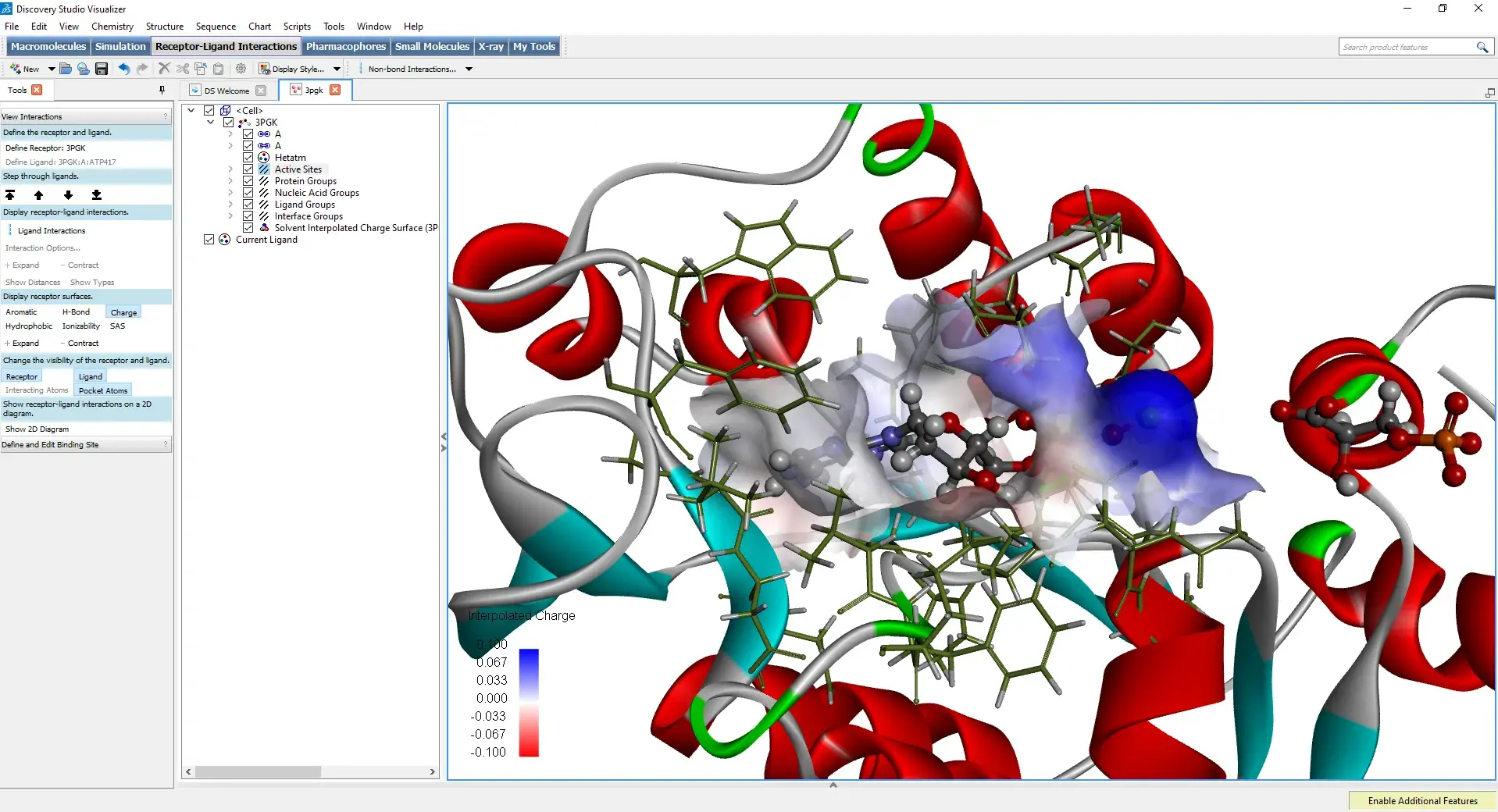
Discovery Studio: The Integrated Suite for Molecular Modeling
What is it?
Dassault Systèmes BIOVIA's Discovery Studio is a comprehensive commercial software suite for molecular modeling and drug design. It offers an extensive range of features, from visualization and preparation of structures to more complex simulations.
- How to use it?
- Advanced structure preparation: He excels in the automated preparation of proteins for docking and simulations, correcting gaps, adding hydrogens, and optimizing side chains.
- Active site analysis: Dedicated tools to identify and characterize binding pockets, calculating their physicochemical properties.
- Docking and virtual screening: He integrates sophisticated docking modules (such as CDOCKER) and virtual screening tools to search for potential ligands in large databases.
- Visualization and publication: Enables the creation of high-quality figures for scientific publications, highlighting ligand-receptor interactions (hydrogen bonds, hydrophobic interactions, pi-stacking, etc.).
- Molecular Dynamics Simulation: To study the dynamic behavior of proteins and ligand-receptor complexes over longer time scales.